Biology is the natural science that studies life and living organisms, including their physical structure, chemical processes, molecular interactions, physiological mechanisms, development and evolution
Tuesday, September 22, 2020
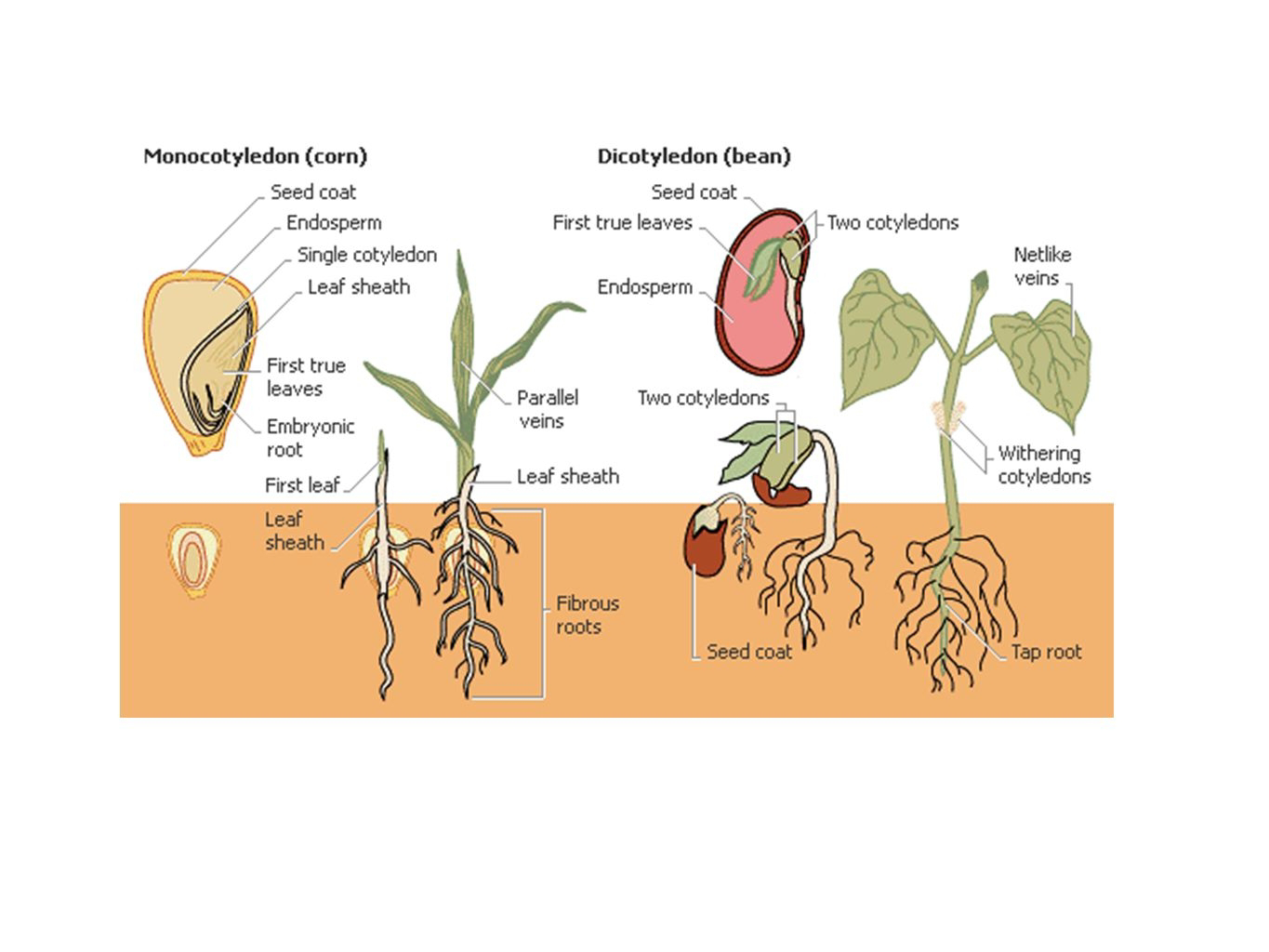
ANGIOSPERMS
Wednesday, September 16, 2020
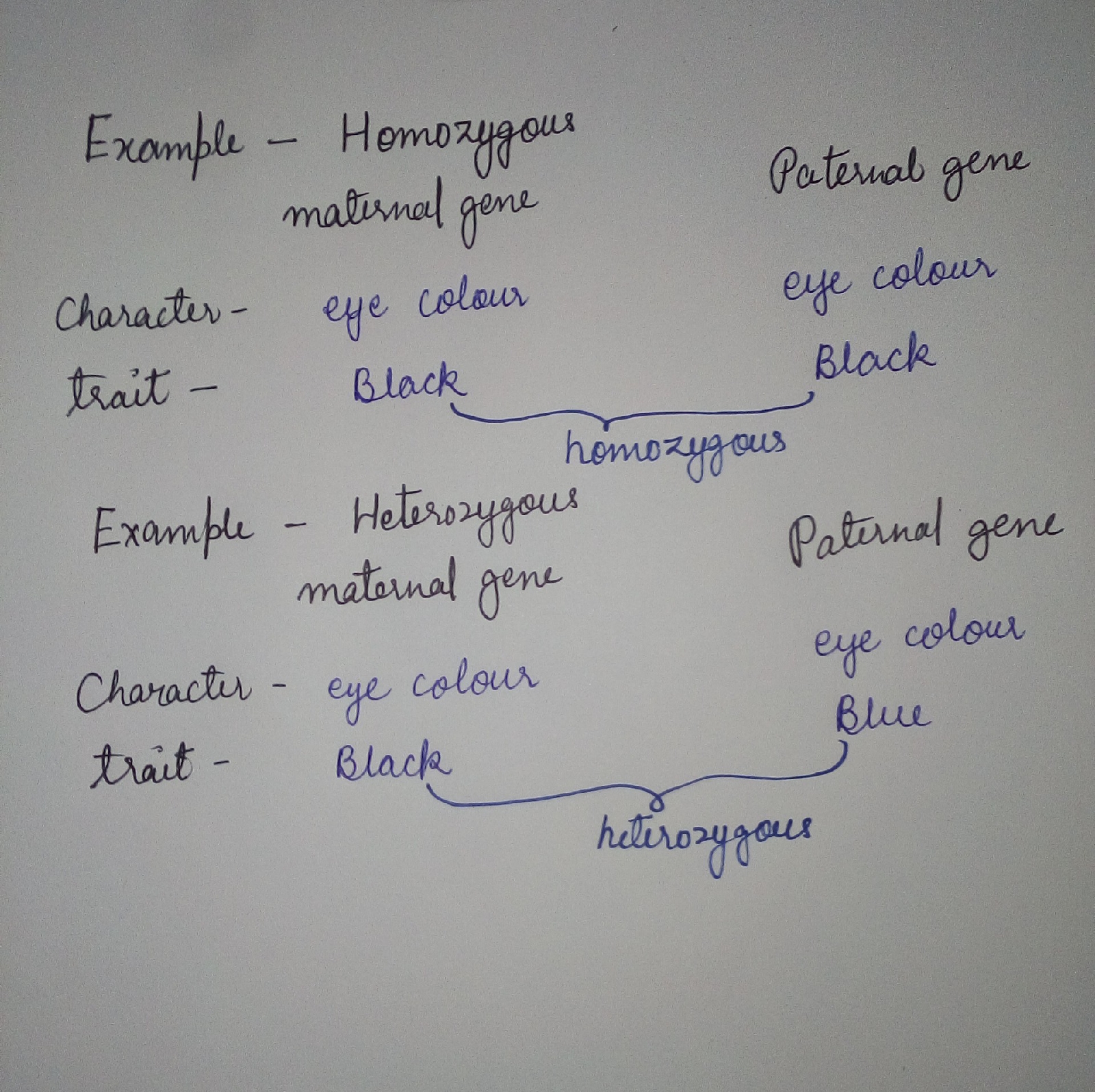
BASIC TERMS OF GENETIC
Monday, September 14, 2020
GENE THERAPY
GENE THERAPY
Gene therapy is when DNA is introduced into a patient to treat a genetic disease. The new DNA usually contains a functioning gene to correct the effects of a disease-causing mutation.In the future, this technique may allow doctors to treat a disorder by inserting a gene into a patient’s cells instead of using drugs or surgery. Researchers are testing several approaches to gene therapy, including:
Replacing a mutated gene that causes disease with a healthy copy of the gene.
Inactivating, or “knocking out,” a mutated gene that is functioning improperly.
Introducing a new gene into the body to help fight a disease.
Although gene therapy is a promising treatment option for a number of diseases (including inherited disorders, some types of cancer, and certain viral infections), the technique remains risky and is still under study to make sure that it will be safe and effective. Gene therapy is currently being tested only for diseases that have no other cures.
TYPES OF GENE THERAPY
There are two different types of gene therapy depending on which types of cells are treated:
- Somatic gene therapy: transfer of a section of DNA to any cell of the body that doesn’t produce sperm or eggs. Effects of gene therapy will not be passed onto the patient’s children.
- Germline gene therapy: transfer of a section of DNA to cells that produce eggs or sperm. Effects of gene therapy will be passed onto the patient’s children and subsequent generations.
GENE THERAPY WORK
Gene therapy is designed to introduce genetic material into cells to compensate for abnormal genes or to make a beneficial protein. If a mutated gene causes a necessary protein to be faulty or missing, gene therapy may be able to introduce a normal copy of the gene to restore the function of the protein.
A gene that is inserted directly into a cell usually does not function. Instead, a carrier called a vector is genetically engineered to deliver the gene. Certain viruses are often used as vectors because they can deliver the new gene by infecting the cell. The viruses are modified so they can't cause disease when used in people. Some types of virus, such as retroviruses, integrate their genetic material (including the new gene) into a chromosome in the human cell. Other viruses, such as adenoviruses, introduce their DNA into the nucleus of the cell, but the DNA is not integrated into a chromosome.
The vector can be injected or given intravenously (by IV) directly into a specific tissue in the body, where it is taken up by individual cells. Alternately, a sample of the patient's cells can be removed and exposed to the vector in a laboratory setting. The cells containing the vector are then returned to the patient. If the treatment is successful, the new gene delivered by the vector will make a functioning protein.
Researchers must overcome many technical challenges before gene therapy will be a practical approach to treating disease. For example, scientists must find better ways to deliver genes and target them to particular cells. They must also ensure that new genes are precisely controlled by the body.
A new gene is inserted directly into a cell. A carrier called a vector is genetically engineered to deliver the gene. An adenovirus introduces the DNA into the nucleus of the cell, but the DNA is not integrated into a chromosome.
IMPORTANCE OF GENE THERAPY
Genes that are flawed and do not work properly can cause disease. Gene therapy is a technique for correcting defective genes responsible for disease development. Researchers may use one of several approaches for correcting faulty genes:
- A normal gene may be inserted into a nonspecific location within the genome to replace a nonfunctional gene. This approach is most common.
- An abnormal gene could be swapped for a normal gene through homologous recombination.
- The abnormal gene could be repaired through selective reverse mutation, which returns the gene to its normal function.
- The regulation (the degree to which a gene is turned on or off) of a particular gene could be altered.
Sunday, September 13, 2020
GENE CLONING AND ITS SIGNIFICANCE
“DNA cloning is a molecular biology technique which is used for the creation of exact copies or clones of a particular gene or DNA.”
The production of exact copies of a particular gene or DNA sequence using genetic engineering techniques is called gene cloning.
- The term “gene cloning,” “DNA cloning,” “molecular cloning,” and “recombinant DNA technology” all refer to same technique.
- When DNA is extracted from an organism, all its genes are obtained. In gene (DNA) cloning a particular gene is copied forming “clones”.
- Cloning is one method used for isolation and amplification of gene of interest.
DNA cloning can be achieved by two different methods:
- Cell based DNA cloning
- Cell-free DNA cloning (PCR)
- DNA fragment containing the desired genes to be cloned.
- Restriction enzymes and ligase enzymes.
- Vectors – to carry, maintain and replicate cloned gene in host cell.
- Host cell– in which recombinant DNA can replicate.
- Steps of gene Cloning:
- The basic 7 steps involved in gene cloning are:
- Isolation of DNA [gene of interest] fragments to be cloned.
- Insertion of isolated DNA into a suitable vector to form recombinant DNA.
- Introduction of recombinant DNA into a suitable organism known as host.
- Selection of transformed host cells and identification of the clone containing the gene of interest.
- Multiplication/Expression of the introduced Gene in the host.
- Isolation of multiple gene copies/Protein expressed by the gene.
- Purification of the isolated gene copy/protein.

- The target DNA or gene to be cloned must be first isolated. A gene of interest is a fragment of gene whose product (a protein, enzyme or a hormone) interests us. For example, gene encoding for the hormone insulin.
- The desired gene may be isolated by using restriction endonuclease (RE) enzyme, which cut DNA at specific recognition nucleotide sequences known as restriction sites towards the inner region (hence endonuclease) producing blunt or sticky ends.
- Sometimes, reverse transcriptase enzyme may also be used which synthesizes complementary DNA strand of the desired gene using its mRNA.
B. Selection of suitable cloning vector
- The vector is a carrier molecule which can carry the gene of interest (GI) into a host, replicate there along with the GI making its multiple copies.
- The cloning vectors are limited to the size of insert that they can carry. Depending on the size and the application of the insert the suitable vector is selected.
- The different types of vectors available for cloning are plasmids, bacteriophages, bacterial artificial chromosomes (BACs), yeast artificial chromosomes (YACs) and mammalian artificial chromosomes (MACs).
- However, the most commonly used cloning vectors include plasmids and bacteriophages (phage λ) beside all the other available vectors.
C. Essential Characteristics of Cloning Vectors
All cloning vectors are carrier DNA molecules. These carrier molecules should have few common features in general such as:
- It must be self-replicating inside host cell.
- It must possess a unique restriction site for RE enzymes.
- Introduction of donor DNA fragment must not interfere with replication property of the vector.
- It must possess some marker gene such that it can be used for later identification of recombinant cell (usually an antibiotic resistance gene that is absent in the host cell).
- They should be easily isolated from host cell.
D. Formation of Recombinant DNA
- The plasmid vector is cut open by the same RE enzyme used for isolation of donor DNA fragment.
- The mixture of donor DNA fragment and plasmid vector are mixed together.
- In the presence of DNA ligase, base pairing of donor DNA fragment and plasmid vector occurs.
- The resulting DNA molecule is a hybrid of two DNA molecules – the GI and the vector. In the terminology of genetics this intermixing of different DNA strands is called recombination.
- Hence, this new hybrid DNA molecule is also called a recombinant DNA molecule and the technology is referred to as the recombinant DNA technology.
E. Transformation of recombinant vector into suitable host
- The recombinant vector is transformed into suitable host cell mostly, a bacterial cell.
- This is done either for one or both of the following reasons:
- To replicate the recombinant DNA molecule in order to get the multiple copies of the GI.
- To allow the expression of the GI such that it produces its needed protein product.
- Some bacteria are naturally transformable; they take up the recombinant vector automatically.
For example: Bacillus, Haemophillus, Helicobacter pylori, which are naturally competent.
- Some other bacteria, on the other hand require the incorporation by artificial methods such as Ca++ ion treatment, electroporation, etc.
F. Isolation of Recombinant Cells
- The transformation process generates a mixed population of transformed and non-trans- formed host cells.
- The selection process involves filtering the transformed host cells only.
- For isolation of recombinant cell from non-recombinant cell, marker gene of plasmid vector is employed.
- For examples, PBR322 plasmid vector contains different marker gene (Ampicillin resistant gene and Tetracycline resistant gene. When pst1 RE is used it knock out Ampicillin resistant gene from the plasmid, so that the recombinant cell become sensitive to Ampicillin.
G. Multiplication of Selected Host Cells
- Once transformed host cells are separated by the screening process; becomes necessary to provide them optimum parameters to grow and multiply.
- In this step the transformed host cells are introduced into fresh culture media .
- At this stage the host cells divide and re-divide along with the replication of the recombinant DNA carried by them.
- If the aim is obtaining numerous copies of GI, then simply replication of the host cell is allowed. But for obtaining the product of interest, favourable conditions must be provided such that the GI in the vector expresses the product of interest.
H. Isolation and Purification of the Product
- The next step involves isolation of the multiplied GI attached with the vector or of the protein encoded by it.
- This is followed by purification of the isolated gene copy/protein.
ITS SIGNIFICANCE
- A particular gene can be isolated and its nucleotide sequence determined
- Control sequences of DNA can be identified & analyzed
- Protein/enzyme/RNA function can be investigated
- Mutations can be identified, e.g. gene defects related to specific diseases Organisms can be ‘engineered’ for specific purposes, e.g. insulin production, insect resistance, etc.
- DNA cloning can be used to make proteins such as insulin with biomedical techniques.
- It is used to develop recombinant versions of the non-functional gene to understand the functioning of the normal gene. This is applied in gene therapies also.
- It helps to analyse the effect of mutation on a particular gene.
Saturday, September 12, 2020
Albinism Disorder
Alkaptonuria Disorder
Thursday, July 30, 2020
ETHICAL ISSUES
Sunday, July 26, 2020
BIOTECHNOLOGY AND ITS APPLICATION
Friday, July 24, 2020
BIOTECHNOLOGY- PRINCIPLES AND PROCESSES
Blog Archive
About
Featured Posts
Featured Posts
Featured Posts
Recent
Featured post
ANGIOSPERMS
ANGIOSPERMS 1. The term "angiosperms" derives from the two Greek words : angeion meaning "vessels" and sperm...